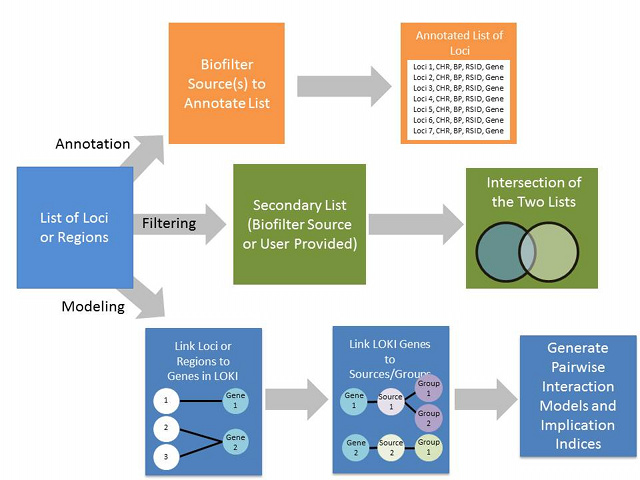
Biofilter is a tool for knowledge-driven multi-SNP analysis of large scale SNP data using information from public databases (The Gene Ontology, The Database of Interacting Proteins, The Protein Families Database, The Kyoto Encyclopedia of Genes and Genomes, Reactome, and Biopath). As part of P-STAR, we will continue to develop the Biofilter resource. In its current implementation, it was primarily developed for common disease GWAS data. There are a number of knowledge sources that would primarily be important to include for pharmacogenomics studies in particular including: PharmGKB VIP genes/pathways, DMET pathways, and eQTL data. We will integrate these and other relevant additional sources of information into the Biofilter.
A network resource for coordination of statistical analysis and methods development in the PGRN.